What we work on
Genomic technology development
CRISPR-based genomic tools for the manipulation of genetically-intractable microorganisms
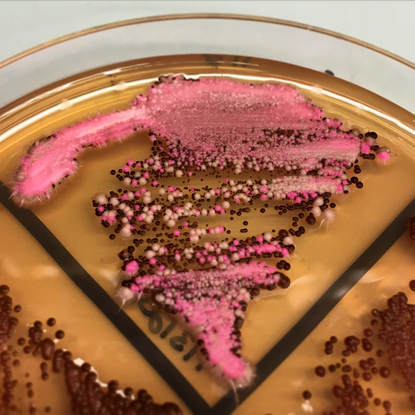
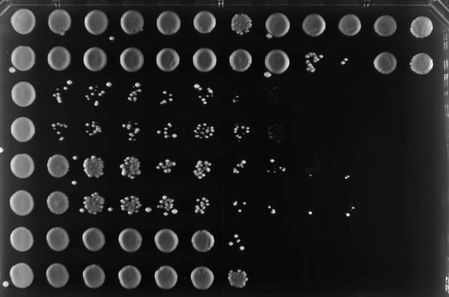
To better study the biology and virulence of fungal pathogens, we are developing new genomic technology platforms for diverse fungal species. We are exploiting CRISPR-Cas9 based technologies to revolutionize the way we do high-throughput functional genomic analysis in fungal pathogens. This is enabling us to map genetic interactions, and uncover genetic factors and pathways that mediate important phenotypes associated with pathogenesis, antifungal drug resistance, and other biological processes.
Relevant publications:
To better study the biology and virulence of fungal pathogens, we are developing new genomic technology platforms for diverse fungal species. We are exploiting CRISPR-Cas9 based technologies to revolutionize the way we do high-throughput functional genomic analysis in fungal pathogens. This is enabling us to map genetic interactions, and uncover genetic factors and pathways that mediate important phenotypes associated with pathogenesis, antifungal drug resistance, and other biological processes.
Relevant publications:
- Development and applications of a CRISPR activation system for facile genetic overexpression in Candida albicans
- Superior conjugative plasmids delivered by bacteria to diverse fungi
- High-content CRISPR screening
- A CRISPR interference system for efficient genetic repression in Candida albicans
- Design, execution, and analysis of CRISPR-Cas9-based deletions and genetic interaction networks in the fungal pathogen Candida albicans
- CRISPR-based genomic tools for the manipulation of genetically-intractable microorganisms
- A CRISPR-Cas9-based gene drive platform for genetic interaction analysis in Candida albicans
- Precise Cas9 targeting enables genomic mutation prevention
Fungal pathogenesis
|
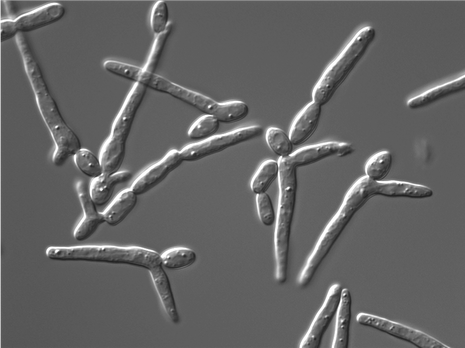
We are interested in microbial fungal pathogens, and the mechanisms by which they cause disease. Most of our work focuses on the human fungal pathogen Candida albicans, but we are also expanding our research to include other fungal pathogens of both humans and plants. We use genetics and systems-level analysis to assess cellular factors involved in fungal pathogenesis and virulence pathways - including cellular morphogenesis and biofilm formation.
Relevant publications:
|
Antifungal drug resistance
|
Resistance to antifungal drugs is a serious clinical concern. We study the mechanisms by which fungal pathogens evolve antifungal drug resistance, and identify genetic factors involved in mediating this resistance. Using experimental evolution, molecular genetic techniques, and whole genome sequencing analysis, we are building an understanding of how fungi evolve resistance to diverse classes of antifungal agents.
Relevant publications:
|